Research Article
Canine Distemper Virus detection based in Hemaglutinine Gene as target in Reverse Transcriptase-Polymerase Chain Reaction

Jara P, Céspedes P and Navarro C*
Department of Animal Preventive Medicine, Faculty of Veterinary Medicine and Animal Sciences. University of Chile, Chile
*Address for Correspondence: Navarro C, Department of Animal Preventive Medicine, Faculty of Veterinary Medicine and Animal Sciences. University of Chile, Chile, Email: [email protected]
Dates: Submitted: 11 December 2018; Approved: 26 December 2018; Published: 27 December 2018
How to cite this article: Jara P, Céspedes P and Navarro C. Canine Distemper Virus detection based in Hemaglutinine Gene as target in Reverse Transcriptase-Polymerase Chain Reaction. Insights Vet Sci. 2018; 2: 034-041. DOI: 10.29328/journal.ivs.1001012
Copyright License: © 2018 Jara P, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Canine Distemper have become a major concern within the veterinary clinical work. Thus, the appearance of many cases of canine distemper in adult animals with their vaccination plan up to date has alarmed veterinarians. Many cases of canine distemper have become a major concern within the clinical veterinary task. The purpose of this work was to detect the gene of hemagglutinin of the virus canine distemper, by using the Polymerase Chain Reaction associated with Reverse transcription (RT-PCR), as confirmation of the clinical diagnosis of disease. For this, peripheral blood samples of animals were used clinically patients and were grouped according to their date of extraction and using vaccines commercial as control.
The results allow to demonstrate a high sensitivity of the technique, besides allowing the use of samples up to seven days of storage at 4°C, despite the fragility of RNA viral.
Detection of canine distemper virus haemagglutinin gene in field samples and its high sensitivity, suggests studying its use as a diagnostic tool complementary to the clinical diagnosis of canine distemper in our country.
Introduction
Etiology
Canine distemper virus (CDV) is taxonomically related to the virus of the measles, the rinderpest virus, the plague virus of small ruminants, the seal distemper virus and the dolphin distemper virus, viruses that are classified within the Mononegavirales order, Paramyxoviridae family, Paramyxovirinae subfamily, Morbillivirus genus [1].
The virus has envelope and a size between 150 to 300 nm in diameter and a chain simple RNA of negative polarity and RNA polymerase [2]. The lipoprotein of the envelope is easily destroyed by lipid solvents, and thus the virus loses its capacity infective It also has the membrane protein (M), two glycoproteins: hemagglutinin (H) and the fusion protein (F) that induce the production of antibodies neutralizing agents, two proteins associated with transcriptases (P and L) and the core protein capsid (N) that covers the viral RNA [1,3,4].
Although there are some antigenic differences between CDV strains shown by serological tests, it is generally accepted that there is only one serotype. But nevertheless, there are considerable differences in the pathogenicity of the different strains isolated [5-7].
Epizootiology
The canine distemper (CD) is enzootic in the world and has a wide range of hosts. Most terrestrial carnivores are susceptible to infection by CDV [5], including the big cats, which are also susceptible to infection and disease by CDV (lions, leopards and tigers in California in 1992 and lions in Tanzania in 1994) [8].
In 1905 Henri Carré discovered the VDC, which causes the most widespread, contagious and lethal multisystem disease of candies and other species of mammals (Mustelidae, Procyonidae, Ursidae, Viverridae, Hyaenidae, Phocidae and Felidae) [9-11].
In Chile, the presence of CD was suggested empirically until 1994 when it was reported the first isolation of the virus in cell cultures inoculated with secretions from a Canine puppy that showed nervous signs with unilateral myoclonus, movements involuntary and ascending paresia of the posterior train. The clinical diagnosis was corroborated by histopathological studies, electron microscopy and direct immunofluorescence [12,13].
The illness
CDV causes a febrile, acute or sub-acute and highly infectious disease in dogs and other carnivores. The overburden cases, characterized by sudden onset of fever and sudden death are rare, but the acute process is frequent. After a period of incubation for three to seven days, animals have a biphasic elevation of temperature, which reaches 41ºC, with a second maximum corresponding to the appearance of a severe leukopenia. During this stage are anorexia, catarrh, conjunctivitis and depression. Some animals show respiratory signs, and others present digestive signs [14,15].
After the onset of the disease, some dogs develop nervous signs after the systemic disease. Depending on the viral strain, these signs can be related to acute illness (attacks and myoclonus with hyperesthesia and depression) or with sub-acute disease (incoordination, ataxia, paresis, paralysis and muscle tremors). Some viral strains produce hyperkeratosis of the footpad and nose [2]. Dogs infected acutely eliminate viruses through all secretions bodily. The most important route of transmission is through aerosols of secretions [5]. This virus removal begins approximately seven days after infection (PI). Dogs that recover after infection with CDV are not immune for life, do not remain persistently infected or eliminate viruses [16].
Diagnosis
There are several techniques for the diagnosis of CD, among them are: histological diagnosis, where inclusion bodies can be found in cells of the oral and conjunctive mucous membranes; the analysis of cerebrospinal fluid, where it is usual dogs with nervous commitment have increased the concentration of proteins and cells mononuclear; or the ELISA test for specific IgM, where antibodies are looked for specific for the virus. However, something that all the tests share is the appearance of false positives, when performed shortly after vaccination of the specimen. In the case of negative results, the absence of the disease cannot be affirmed either, since both in the early stages of the infection and in very late these tests they show a very low sensitivity, so they are only diagnostic approximations. Among the definitive diagnoses are viral isolation, which does not constitute a routine technique within the diagnostic laboratories and the RT-PCR technique, which allows to detect more sensitively the presence of the virus [5,17]. Currently, the diagnosis of CD in Chile continues mainly in the hands of the clinical signology, for which no studies have yet been reported to detect differences between national CDV isolates at the genomic, antigenic or biological level that could explain something troubling: the presentation of the clinical picture in dogs with their daily vaccination program.
Vaccines
CDV is characterized by damaging innate and adaptive immunity from moments of the infectious disease due to its high lymphotropism and ability to generate change of essential functions in immune cells. It has been described that attenuated viruses currently used in modified live vaccines maintain their lymphotropism and ability to induce immunosuppression, compromising the balance of responses previously mentioned [18,19].
In Chile, it is advisable to vaccinate with monovalent vaccine between ten and twelve weeks; to fourteen or sixteen weeks and twenty-four weeks. When using the sixfold vaccine (CDV, leptospira, hepatitis 1 and 2 viruses, canine parvovirus type 2 and parainfluenza type 2) the scheme advises to apply the parvovirus vaccine at eight weeks; at ten weeks sixfold vaccine and at sixteen weeks the sixfold vaccine. The recombinant vaccine eightfold applies from six weeks of age and every twenty-one days until twelve weeks (three doses) [20].
In this report, the general objective was to detect the gene of the hemagglutinin (H) of CDV by RT-PCR, as confirmation to the clinical diagnosis of the disease.
1. For this purpose, it was proposed: to detect the gene of Hemagglutinin in commercial vaccines NOBIVAC® PUPPY and CANIGEN MHA2PPi / L for CDV by RT-PCR, used as reference strains
2. Detect the gene of the CDV hemagglutinin obtained from suspicious clinical samples.
Materials and Methods
Design
The experimental design used in this work included the detection of the CDV H gene in fresh blood samples (one week of obtained) and samples with one or more months of obtained.
Samples: Thirty samples were taken consisting of 2 mL of peripheral blood from dogs of any age and sex, suspected of being in the disease and positive to the test of IgM specific for CDV. For this purpose, it was supported by the Santiago Veterinarian Laboratory, the Neurological Veterinary Institute and the Vet-lab® laboratory. Samples with and without anticoagulant were refrigerated at 4° C for later extraction.
As a positive control, two commercial CANIGEN vaccines (CCV) were used MHA2PPi / L (103-105 TCID50) and NOBIVAC® PUPPY (103-105 TCID50) that contain the Lederle and Onderstepoort strains.
Detection of the H gene in CCV. Sensitivity by the technique
For this purpose, the CCV was used MHA2PPi / L (103-105 TCID50), which was reconstituted in 1 ml of nuclease-free water and prepared dilutions to the tenth (10-1 to 10-7), that is from a range between 50 and 0.5 TCID50 up to an interval between 0.005 and 0.00005 TCID50 to obtain an approximation to the sensitivity of the diagnostic test against the old CDV strains.
Samples of one or more months old
Twenty samples that were processed were grouped into four groups with 5 samples each: samples with anticoagulant one month old, samples without anticoagulant a month old, samples with anticoagulant of two or more months old and samples without anticoagulant of two or more months old Thus, the samples contained in tubes with anticoagulant were homogenized gently, an aliquot of 250 uL was taken and separated into tubes, then all samples were centrifuged at 200g for ten minutes. Serum or plasma and its phlogistic layer. Aliquots of 250 uL were prepared and RNA was extracted.
Samples one week old
Eight samples were processed corresponding to one-week old clinical samples stored in tubes with anticoagulant and two commercial vaccines. Thus, the samples were homogenized gently, an aliquot of 250 uL was taken and separated into tubes, then the Histopaque © 1077, Sigma-Aldrich® product to obtain the phlogistic layer of the shows and thus get higher viral concentration to proceed to the detection of the virus. 250 uL aliquots were prepared and RNA extraction was carried out. In parallel, two samples one week old, different from the previous ones, stored in tubes with anticoagulants, positive against the RT-PCR kit used by Vet-lab®, were processed to make the comparison in the detection sensitivity from the direct sample and a show following the protocol for the extraction of the phlogistic layer.
Viral RNA extraction
The viral RNA was obtained by means of an extraction kit (Trizol LS, Invitrogen ©). Briefly, 250 uL of sample were mixed with 750uL of Trizol reagent. It was left to ambient temperature (AT) for five minutes and subsequently 0.2 mL of chloroform. They were mixed vigorously for fifteen seconds and left to rest at AT for five minutes. Subsequently, it was centrifuged at 7000g for fifteen minutes, the aqueous phase was transferred to a clean tube and a volume of isopropanol was added. It was left at AT for ten minutes and centrifuged at 7000g for ten minutes. Then the supernatant was removed, washed three times with ethanol (1 mL, 75% ethanol), vortexed for fifteen seconds and centrifuged at 2000g for five minutes. The supernatant was removed, the precipitate dried under vacuum for ten minutes and re suspended in 100uL of sterile water, free of nucleases finally, the RNA was incubated at 55-60 ° C for ten minutes and stored at -20° C.
RT-PCR
The region to be amplified is the hemagglutinin gene of the CDV. The CDH13 primers were used: 5'-CAAGACAAGGTGGGTGCCTT-3' and CDVH18: 5'-CTTGG TGAAATCG AACTCCA-3', which adhere between nucleotide 7091-7110 and between 7265-7246 respectively, which generate a DNA fragment of 174 base pairs (bp) [21].
Reaction mixture
In the RT-PCR reaction the "SuperScript one step RT-PCR with platinum Taq" kit was used (Invitrogen®) following the protocol proposed by the company, which consisted of 25 uL of the "2x Reaction Mix" component, which contains 0.4 mM of each deoxyribonucleotide and 3.2 mM of MgSO4, 2uL of "SuperScript. III RT / Platinum Taq Mix ", 2uL of each splitter and 19 uL of RNA tempering to reach a final volume of 50uL.
RT-PCR Protocol
The protocol contemplated an incubation at 50° C for forty minutes, then at 94° C for two minutes, to then carry out thirty-five cycles (94° C for one minute; 55° C for two minutes; 72ºC for two minutes) and a final extension at 72ºC for two minutes [22]. The primers were commissioned to Bioscan® for its synthesis.
Visualization of amplified products
The visualization of the amplified product was carried out by gel electrophoresis of 2% agarose (Winkler®) in Tris acetate EDTA (TAE) buffer (Fermentas®) and later staining with ethidium bromide (0.5 μg / ml) (Fermelo®) for thirty minutes.
Results
Detection of the H gene in CCV. Sensitivity by the technique
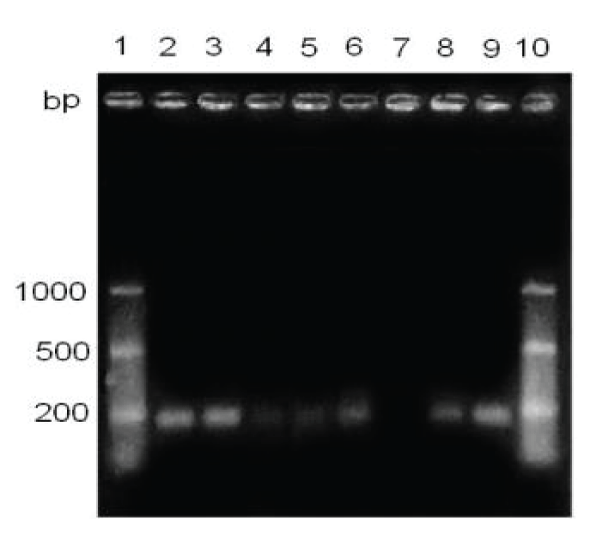
Two bands were visualized corresponding to a fragment of DNA of approximately 174 bp, in the lanes of both commercial vaccines (Figure 1).
Detection of the H gene of canine distemper virus in samples stored with a week old
It is possible to visualize a fragment of approximately 174 bp, both in the positive controls and in five of six clinical samples (Figure 1).
Effect of the staining time of the agarose gel on the detection sensitivity of the RT-PCR technique
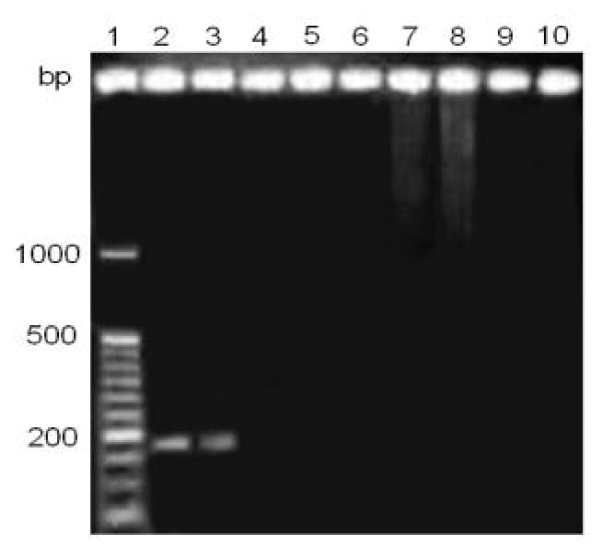
By studying the effect of staining time on the sensitivity of detection of the RT-PCR, it was determined that 60 minutes of staining could be detected 5/6 positive samples to CDV, however, with 30 minutes of staining, all six samples clinical studies were negative (Figures 1,2).
Figure 1: Detection of the hemagglutinin gene in clinical samples, with 60 minutes of staining. Lane 1 and 10, marker of molecular size from 50 to 1000 bp .; lane 2, CANIGEN vaccine MHA2PPi / L; lane 3, vaccine NOBIVAC® PUPPY; lanes 4-9, clinical samples, with anticoagulant, one week old.
Figure 2: Detection of the hemagglutinin gene in clinical samples, with 30 minutes of staining. Lane 1, marker of molecular size from 50 to 1000 bp .; lane 2, CANIGEN vaccine MHA2PPi / L; lane 3, NOBIVAC® vaccine PUPPY; Lane 4-10, clinical samples, with anticoagulant, one week old.
Approach to the sensitivity of the technique
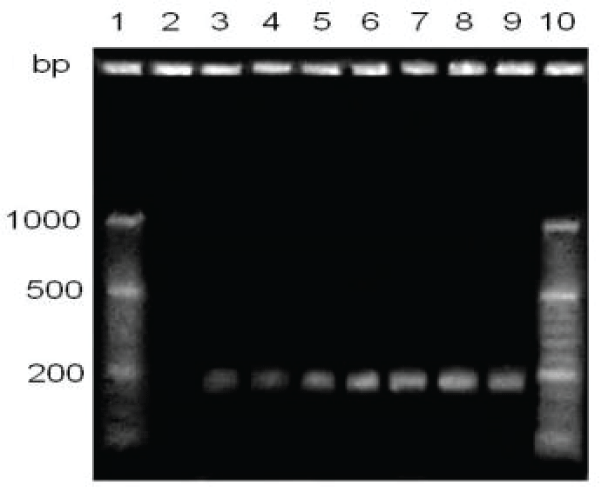
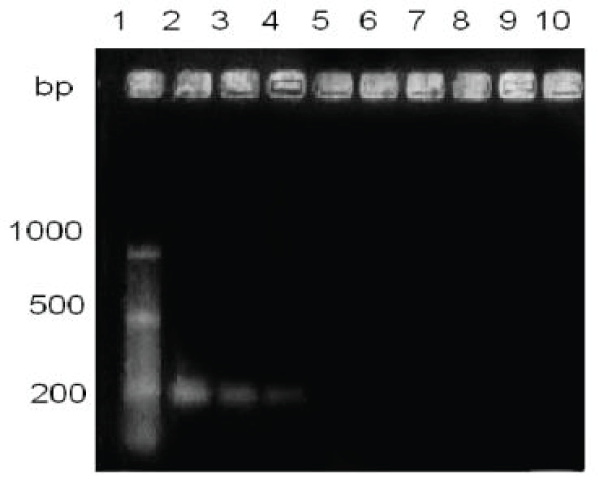
By making seven dilutions to the tenth from 10-1 up to 10-7 from the commercial vaccine CANIGEN MHA2PPi / L (103-105 TCID50), and a sixty minutes ethidium bromide stain bands were visualized up to one dilution of 10-6 TCID50, that is between 0.0019 and 0.000019 TCID50 (Figures 3,4).
Figure 3: Sensitivity comparison of hemagglutinin gene detection, with 60 minutes staining. Lane1, molecular size marker; lanes 2 and 4, direct clinical sample; lanes 3 and 5, samples processed with protocol Histopaque (comparison between 2/3 and 4/5); lanes 6-9, CANIGEN vaccine MHA2PPi / L diluted from 10-1 to 10-4.
Figure 4: Comparison of sensitivity in the detection of the hemagglutinin gene, with a 60-minute stain. Lane1, molecular size marker; lane 2, CANIGEN vaccine MHA2PPi / L; lanes 3 and 4, CANIGEN vaccine MHA2PPi / L diluted from 10-5 to 10-6.
Use of Histopaque © 1077 (Sigma-Aldrich®). By studying the effect of the influence of Histopaque © 1077 on the sensitivity of the technique, it was observed that both samples. Clinics extracted with Histopaque © 1077 were positive by RT-PCR, and only one of they were positive by extraction from the direct sample (Figure 3).
Detection of the H gene of canine distemper virus in samples stored with a month or more than seniority
The 20 samples did not present visible bands in the transilluminator of UV light. Only in the case of positive controls (commercial vaccines) was the visualization of a DNA fragment of approximately 174 bp (the image of the results was not attached at work).
Discussion
The cases of CD are a major concern within the task veterinary clinic. Thus, in recent years, the appearance of many cases has alarmed the veterinary doctors, since they have a special characteristic, the appearance of the disease in adult animals and with its vaccination plan up to date. This leads to the search for a CD confirmatory diagnostic method and that allow timely treatment of the disease [21]. Worldwide CD cases have been associated with a poorly performed vaccination plan, or to the suspicion of a residual virulence in vaccines [23]. The RT-PCR technique for the detection of the hemagglutinin gene was selected because this protein expressed by the virus is responsible for generating the response immune in the organism, besides being used worldwide for its diagnosis [24]. The hemagglutinin gene has the greatest variation, reaching 10%, being used generally for the establishment of phylogenetic trees of the virus [25-27].
In this report, a fragment of 174 bp was amplified in both clinical samples as in vaccine strains, which would allow its use as a diagnostic tool confirmation of the clinical diagnosis, as an alternative to the diagnostic kit of the company BioingenTech®, currently used, regardless of whether this kit appears to be is detecting a different gene: the N gene [28]. The results suggest a high sensitivity of the technique implemented in this report that would allow the use of blood samples up to seven days after being extracted from the animal, despite the fragility of viral RNA [14]. In relation to the detection of the 174 bp fragment in commercial vaccines, it was possible to perform an estimate of the sensitivity of the technique and the results show a high sensitivity. RT-PCR designed for the same gene segment, reach up to detect 103 copies of the viral RNA, however the strategy used in this report prevent compare both values [1].
The use of Histopaque© 1077 reagent (Sigma-Aldrich®), was able to increase the sensitivity of the technique, becoming a recommended step within the protocol. This is because it allows to separate the phlogistic layer of a blood sample, where the cells of the white line, white of infection of the CDV, are located, thus obtaining a higher viral concentration.
One of the critical points that must be considered during the process of the technique is the incubation time in ethidium bromide. In this report, it was established that increase the staining time of the genomic material to double (60 minutes), it could be significantly increase the sensitivity of the technique. As this reagent is of character carcinogenic, an alternative to study is the use of an analogous product like GelRed Nucleic Acid Gel Stain TM. However, the effect on sensitivity should be analyzed of the technique.
Although, there are variations in the H gene represented by amino acid substitutions in key epitopes of protein H, apparently the 174 bp fragment is a good choice as target of detection in clinical samples and probably visualization of the DNA bands can be improved by increasing from 35 to 40 cycles in the of the PCR, modifying the protocol used [22].
Conclusion
This work allowed the molecular detection of CD and represented the first step towards the molecular characterization of CDV in Chile [29], detection in maned wolfs [30] and the use of other genes for the detection of the virus [31].
References
- Martella V, Elia G, Lucente MS, Decaro N, Lorusso E, et al. Genotyping canine distemper virus (CDV) by a hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet Microbiol. 2007; 122: 32–42. Ref.: https://goo.gl/Za8yvN
- Fenner F, Bachmann PA, Gibbs EP, Murphy FA, Studert MJ, et al. Veterinary virology. 349-386. Ref.: https://goo.gl/RRaa9u
- Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JP, et al. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem.2012; 287: 16324-16334. Ref.: https://goo.gl/zKkS5i
- Avila M, Alves L, Khosravi M, Ader N, Origgi F, et al. Molecular determinants defining the triggering range of prefusion F complexes of canine distemper virus. J Virol. 2014; 88: 2951-2966. Ref.: https://goo.gl/Nv4oEa
- Appel MJ, Summers BA. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet. Microbiol. 1995; 44: 187–191. Ref.: https://goo.gl/3FJu5n
- Tan B1, Wen YJ, Wang FX, Zhang SQ, Wang XD, et al. Pathogenesis and phylogenetic analyses of canine distemper virus strain ZJ7 isolate from domestic dogs in China. Virol J. 2011; 8: 520. Ref.: https://goo.gl/8uJscR
- Terio KA, Craft ME. Canine Distemper Virus (CDV) in Another Big Cat: Should CDV Be Renamed Carnivore Distemper Virus? mBio. 2013; 4: e00702-13 Ref.: https://goo.gl/6TvADv
- Appel MJ, Yates RA, Foley GL, Bernstein JJ, Santinelli S, et al. Canine distemper virus epizootic in lions, tigers and leopards in North America. J Vet Diagn Invest. 1994; 6: 277–288. Ref.: https://goo.gl/4bfuwh
- Deem SL, Spelman LH, Yates RA, Montali RJ. Canine distemper in terrestrial carnivores: a review. J. Zoo. Wildl. Med. 2000; 31: 441-451. Ref.: https://goo.gl/nK6ZD7
- Pardo ID, Johnson GC, Kleiboeker SB. Phylogenetic characterization of canine distemper viruses detected in naturally infected dogs in North America. J. Clin. Microbiol. 2005; 43: 5009-5017. Ref.: https://goo.gl/wHP6vK
- Lamb R, Parks G. Paramixoviridae: the viruses and their replication.In: Knipe, D.; Howley, P. Fields Virology 5ª ed. Lippincott Williams & Willkins. Philadelphia, USA. 2007; 1449-1496. Ref.: https://goo.gl/JuHA92
- Cerda L, Mathieu C, Quinteros G. Primer aislamiento de virus distemper canino en Chile. XIV Congreso Panamericano de Ciencias Veterinarias, Acapulco, México. 1994;
- Navarro C, Pizarro J, Celedón M. Virus distemper canino en Chile. XII Congreso Nacional de Medicina Veterinaria, Chillán, Chile. 2002;
- Murphy F, Gibbs E, Horzinek M, Studdert M. Veterinary Virology: The Third Edition. Elsevier. California, USA. 1999; 629. Ref.: https://goo.gl/jz3FD4
- Carvalho OV, Botelho CV, Torres CG, Scherer PO, Pinheiro JA, et al. Immunopathogenic and neurological mechanisms of canine distemper virus. Adv Virol. 2012; 2012: 163860. Ref.: https://goo.gl/C1AoHo
- Perrone D, Bender S, Niewiesk S. A comparison of the immune responses of dogs exposed to canine distemper virus (CDV) - Differences between vaccinated and wild-type virus exposed dogs. Can J Vet Res.2010: 74: 214-217. Ref.: https://goo.gl/cS6pZA
- Cho HS, Park NY. Detection of canine distemper virus in blood samples by reverse transcription loop-mediated isothermal amplification. J Vet Med B Infect Dis Vet Public Health. 2005; 52: 410-413. Ref.: https://goo.gl/cmhsst
- Sereda AD, Gavrilov KE, Fugina LG. Distemper of carnivore: proliferative activity of lymphocytes in sick and vaccinated dogs. Vopr Virusol. 1999; 44: 257-261. Ref.: https://goo.gl/jf6NBe
- Céspedes PF, Cruz P, Navarro C. Modulación de la respuesta inmune durante la infección por virus distemper canino: implicancias terapéuticas y en el desarrollo de vacunas. Arch Med Vet. 2010; 42: 15-28. Ref.: https://goo.gl/6mLCju
- Berríos P, Durán C. Principales enfermedades virales de los caninos. Situación en Chile. Monogr Electron Patol Vet. 2005; 2: 68-93. Ref.: https://goo.gl/2qbQ84
- Iwatsuki K, Miyashita N, Yoshida E, Gemma T, Shin Y, et al. Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. Journal of General Virology. 1997; 78: 373–380. Ref.: https://goo.gl/82ECTm
- Mochizuki M, Hashimoto M, Hagiwara S, Yoshida Y, Ishiguro S. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J Clin Microbiol. 1999; 37: 2936-2942. Ref.: https://goo.gl/eLNKWK
- Martella V, Cirone F, Elia G, Lorusso E, Decaro N, et al. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Veterinary Microbiology. 2006; 116: 301–309. Ref.: https://goo.gl/ktwa8f
- Gemma T, Miyashita N, Shin YS, Okita M, Mori T, et al. Serological survey of canine distemper virus infection using enzyme-linked immunosorbent assay. J Vet Med Sci. 1995; 57: 761-763. Ref.: https://goo.gl/72s5wz
- Blixenkrone-Möller M, Svansson V, Appel M, Krogsrud J, Have P, et al. Antigenic relationship between field isolates of morbilliviruses from different carnivores. Arch. Virol. 1992; 123: 279–294. Ref.: https://goo.gl/5AEzh4
- von Messling V, Zimmer G, Herrler G, Haas L, Cattaneo R. The Hemagglutinin of Canine Distemper Virus Determines Tropism and Cytopathogenicity. J Virol. 2001; 75: 6418-6427. Ref.: https://goo.gl/jRyFu4
- Ke GM, Ho CH, Chiang MJ, Sanno-Duanda B, Chung CS, et al. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet Res. 2015; 11: 164. Ref.: https://goo.gl/yzJziv
- Shin Y, Mori T, Okita M, Gemma T, Kai C, et al. Detection of canine distemper virus nucleocapsid protein gene in canine peripheral blood mononuclear cells by RT-PCR. J Vet Med Sci.1995; 57: 439–445. Ref.: https://goo.gl/cDdWQz
- Salas V, Pizarro J, Navarro C. Phylogenetic analysis of canine distemper virus detected in Chile. International Journal of Current Research. 2018; 10: 72402-72407. Ref.: https://goo.gl/rggJ2k
- Abarca MJ, Hidalgo E, Raggi LA, Navarro C. Genotypic evidence of infection by Canine distemper virus in maned wolf from a zoological collection in Chile. IJSER. 2018; 9: 2055-2064. Ref.: https://goo.gl/mf3u4v
- Gallegos M, Céspedes P, Pizarro J, Navarro C. Is the M gene of Canine Distemper virus an eligible target for detection? East African Scholars J Agri Life Sci. 2018; 1: 22-32. Ref.: https://goo.gl/dAjV9U
- Harder TC, Kenter M, Vos H, Siebelink K, Huisman W, et al. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J Gen Virol. 1996; 77: 397–405. Ref.: https://goo.gl/53K3iz