Research Article
Use of the Reverse Transcription-Polymerase Chain Reaction for differential detection of two lineages of the canine distemper virus in Chile

Bolívar P1, Céspedes PF2 and Navarro C1*
1Animal Preventive Medicine Department, Faculty of Veterinary and Animal Sciences, FAVET, University of Chile, Chile
2The Kennedy Institute of Rheumatology. University of Oxford, UK
*Address for Correspondence: Navarro C, Animal Preventive Medicine Department, Faculty of Veterinary and Animal Sciences, FAVET, University of Chile, Chile, Email: [email protected]; [email protected]
Dates: Submitted: 18 February 2019; Approved: 01 March 2019; Published: 04 March 2019
How to cite this article: Bolívar P, Céspedes PF, Navarro C. Use of the Reverse Transcription-Polymerase Chain Reaction for differential detection of two lineages of the canine distemper virus in Chile. Insights Vet Sci. 2019; 3: 005-013. DOI: 10.29328/journal.ivs.1001014
Copyright License: © 2019 Bolívar P, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: RT-PCR; Hemagglutinin; H gene; America-1; Europe-1/South America-1; Canine distemper virus.
Abstract
Worldwide, Canine Distemper Virus (CDV) infection is a highly prevalent disease with high morbidity and mortality. CDV causes a multisystemic disease in a wide range of hosts including 9 families of mammals among them some primates, cetaceans and numerous carnivores. It presents a high tropism for lymphoid, neurological and epithelial tissue, leading to an infection of almost all systems, so the clinical signs observed are very varied. The diagnosis is made based on the clinical presentation of the disease, which considers a variety of signs and must be confirmed by a laboratory diagnostic method. The molecular technique called Reverse Transcription Polymerase Chain Reaction (RT-PCR) has been used to characterize viral strains based on the basis of genetic differences on the hemagglutinin (H) gene of CDV has allowed the identification of 14 circulating lineages in the world. Two lineages, namely the America-1 and the Europe-1/South America-1 have been described in Chile. The goal of this work was to implement a multiplex RT-PCR protocol, which was built on the in silico design of primers based on the H gene nucleotide sequences stored in the Genbank® database. This method was capable of detecting the previously described two circulating genetic lineages of CDV in a differential way providing a supporting diagnostic tool for epidemiological studies in the country. These results suggest that the primers described here are extremely selective for the above-mentioned lineages. In addition, our initial screening indicated that most analyzed clinical samples corresponded to the America-1 lineage, stressing the need for a continuous surveillance in order to properly address the prevalence of both lineages in Chile.
Introduction
The Canine Distemper Virus (CDV) infection is one of the most severe infectious diseases in dogs and other carnivores, being multi-systemic, highly contagious and with a worldwide burden. The pathology caused by CDV has a variable morbidity and mortality according to the species, which can exceed 50% in dogs and over 90% in ferrets. As a single-stranded and negative sense RNA genome virus CDV has been classified within the Mononegavirales order, the Paramyxoviridae family and the Morbillivirus genus. Remarkably, despite CDV has a single antigenic type (i.e. serotype), to date more than 14 different lineages have been described worldwide. Lineage determination has been based mostly in genes encoding viral glycoproteins, of which the analysis of the hemagglutinin (H) gene is one of the most informative given its high genetic variability.
CDV presents a high tropism for lymphoid, neurological and epithelial tissue, which leads to a multisystemic infection affecting among others the digestive, respiratory, and central nervous systems. Therefore, clinical manifestations associated with the disease are broad and include most frequently hyperqueratosis of nose and footpads, mucopurulent respiratory secretions, dyspnea, generalized lymphoadenopathy, diarrhea, myoclonus, and seizures. Due to this high number of clinical manifestations, it is necessary to have laboratory diagnostic tools that allow confirming the presence of the virus. Currently, there are several techniques such as immunofluorescence, ELISA, RT-PCR, among others, to fulfill this purpose. The RT-PCR has promoted the identification of the circulating viral lineages, such as the two characterized in Chile namely, América-1 and Europa-1 / Sudamérica-1.
Thus, we developed an RT-PCR technique capable of both detecting and differentiating these two prevalent lineages by designing primers based on official nucleotide sequences deposited in the Genbank®. In this way, we would be capable of characterizing a clinical viral strain, to corroborate its genetic lineage and to assess whether other CDV lineages are circulating in the country in case of negative detection. The latter alternative would open the option that another Canine Distemper Virus lineage, not yet determined, be present in the country.
Etiology of the canine distemper virus
CDV is the etiological agent that causes the CD. It belongs to the genus Morbillivirus, family Paramixoviridae, and order Mononegavirales. As an enveloped virus containing a lipid membrane enriched in glycoproteins needed for the infection of host cells, CDV is a very labile in the environment and susceptible to most disinfectants. It contains a non-segmented RNA genome of negative polarity and has six genes that code for the six structural proteins present in infective viral particles (virions): the matrix protein (M); the hemagglutinin (H) and fusion protein (F), which are two glycoproteins involved in binding and infection of target cells. Also, CDV encodes three proteins involved in the replication of the viral genome and the transcription of viral proteins upon infection of target cells: the phosphoprotein (P), the large RNA-dependent RNA polymerase (L) and the nucleocapsid protein (N), which wraps the viral genome to form ribonucleoprotein complexes [1-3].
Lineages
The H gene, which encodes the surface glycoprotein hemagglutinin has the most genetic heterogeneity within species of the Morbillivirus genus, and hence, is sequencing is being used in the characterization of viral strains. Within the CDV, the amino acid variation of hemagglutinin reaches 10% within the strains. This variability, together with the phylogenetic analysis of this H protein, has allowed the definition of different CDV genetic lineages circulating globally [4]. To date, 14 circulating lineages of the CDV have been described: Africa-1; Africa-2; America-1; America-2; Asia-1; Asia-2; Asia-3; Asia-4; Europe-1 / South America-1; Europe-2 (wild European); Europe-3 (Arctic); Rockborn-like; South America-2; South America-3 [5-8].
Epidemiology
The CDV has a wide range of hosts where it is pathogenic. Among them, we can mention numerous families of the order Carnivora, such as Canidae (dogs and foxes), Felidae (wild cats), Mustelidae (ferrets and minks), as well as the Pinnipedia superfamily, some cetaceans and Japanese primates, which would cause a Serious threat in the conservation of globally endangered species. Worldwide, the disease is highly prevalent having also a high morbidity and mortality, which varies broadly among carnivores [9-11].
In Chile, CD outbreaks have officially been reported in foxes and dogs of the Coquimbo region and Robinson Crusoe Island, respectively. The latter case generated great concern about the potential risk of contagion to the two-headed wolf of Juan Fernández (Arctocephalus philippii) a protected, endangered species in the country [12-14].
Pathogenesis
The CDV is highly contagious and can enter a susceptible host by nasal or oral route, from oronasal secretions or urine from an infected animal. Within hours of infection, CDV targets immune cells expressing surface CD150 to initiate its replication, such as alveolar macrophages and / or dendritic cells of the respiratory tract. These infected cells spread the virus through the lymphatic system and blood, establishing a primary, cell-associated viremia between 3 and 6 days after the infection. By 10 days post-infection, a subsequent viremia leads CDV dissemination towards the epithelial tissues of several systems, including immune, neurological and epithelial tissues, which causes a wide variety of clinical signs associated to a loss of function of the affected systems [11,15,16]. Importantly, after the secondary viremia and if the host antiviral immune response fails in controlling CDV dissemination, the virus reaches central nervous system (CNS), causing a neurologic presentation of the disease. Less frequently, disorders of the urogenital, cardiac and dermatological system are also observed [11]. CDV spreads in all secretions and excretions after 5 days after infection, before the onset of clinical signs. Dissemination can continue for up to 3 to 4 months, but usually lasts 1 to 2 weeks [15].
Clinical manifestations of the disease
The clinical signs of distemper in canines are very varied. Its severity, duration and presentation depend on the age of the animal, its immune status and the strain’s virulence, as well as concomitant infections with other pathogens. In the first instance, the animal will attend with lymphopenia, transient fever and deep immunosuppression. During the second phase of viremia, the various clinical manifestations will be presented according to the affected tissue. However, the first clinical signs are characterized by lethargy, dehydration, anorexia and weight loss [11,15,17,].
Dogs with respiratory compromise may present bilateral nasal serous to mucopurulent discharge, conjunctivitis and non-productive cough. Enteric infection leads to epithelial destruction of the gastrointestinal tract and thus, inappetence, vomiting, diarrhea, electrolyte abnormalities, which accompany the observed dehydration [11,15].
The neurological signs are progressive and will depend on the distribution of the virus within the CNS and include hyperesthesia, myoclonus, seizures, nystagmus, ataxia, paralysis or postural deficit, among others. In a more chronic way, dermatological and ocular manifestations can be observed, such as pustular dermatitis, hyperkeratosis of the plantar pads and nasal epithelium, uveitis, chorioretinitis and dry keratoconjunctivitis [11,15,17,].
Vaccination
Vaccination is the main strategy to control the disease, for which vaccines with live attenuated viruses and recombinant vaccines have been developed. Immunization has considerably reduced the incidence of CDV in the population of domestic dogs, however, outbreaks of the disease have been observed in vaccinated dogs around the world. Although the causes of these events have not been determined, the outbreaks could be explained by an insufficient attenuation of the vaccine virus, by human errors (incorrect vaccination plans, inadequate administration of vaccines, poor storage or handling of these), or by genetic variations of the virus (strains capable of evading the immune response induced by vaccines) [5,6,15,18,].
Diagnosis
The definitive diagnosis of CDV infections should be based on the clinical presentation of the disease and the identification of signs associated with the involvement of multiple systems and a history of risk, such as exposure to infected dogs, lack of vaccination or the improper vaccination of dogs with co-morbidities [15,19].
However, it is necessary to establish a laboratory diagnostic method to confirm the presence of the virus and exclude other diseases with similar clinical manifestations [20]. Some diagnostic techniques include viral isolation, direct immunofluorescence, ELISA for the detection of specific IgM against CDV and the Reverse Transcription-Polymerase Chain Reaction (RT-PCR) [15,17].
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
The RT-PCR technique allows the study of viral genetic variations, through the molecular characterization of genome sequences. Thanks to this technique, it has been possible to identify different lineages circulating worldwide and in Chile, mostly through the analysis of the H gene of the virus [17]. In this context, a multiple RT-PCR protocol was developed using primers designed in silico capable of detecting and differentiating the America-1 and Europe-1/South America-1 lineages. Thus, we sought to create a diagnostic tool also useful for epidemiological studies informing about the prevalence of both lineages in the country. This method will also allow us to evaluate the presence of additional lineages as whenever a known positive sample by other means appears negative after our analyses will suggest that another circulating lineage of CDV, not yet described, exists in the country.
Materials and Methods
The present report was carried out in the Virology and Microbiology laboratories of the Department of Preventive Animal Medicine at the School of Veterinary and Animal Sciences of the Universidad de Chile.
Primers design
For the design of the primers of both lineages, a phylogenetic tree of the H gene of CDV described by Ke et al. [21], was used, using the access numbers for the 11 official nucleotide sequences that make up the lineage. América-1 and for the 34 official nucleotide sequences of the Europe-1/South America-1 lineage, stored in the Genbank database (Annex 1). Collected sequences were then analyzed for nucleotide identity using the Clustal Ω online program to obtain the conserved consensus regions that were used in the design of the primers for each lineage. This design was made using the free online program Primer-BLAST of the National Center for Biotechnology Information, considering certain characteristics that the partisans had to possess, which were: to have a percentage of cytosine and guanine bases close to 50%; that the amplicon fragment obtained by each pair of primers was of different size; that the difference in melting temperature for each pair was not greater than 3° C; and that this melting temperature was similar for both pairs of primers.
After obtaining the ideal pair of primers for each lineage, their sequences were compared with the other component sequences of each lineage of the CDV H gene. In addition, their sequences were surveyed against other sequences by BLAST online program, where the specificity of each pair of primers could be determined. The sequences of the primers were sent to be synthesized at the company Fermelo® and corresponded to P1: 5'-CTATTG CATCGGCAGCAAATC-3’ and to P2: 5'-CCCAGTGTCACTACTAGAATA CC-3' for the detection of the America-1 lineage, primers that allowed the amplification of a DNA fragment of around 478 base pairs (bp) (Annex 2). While the sequences synthesized for the detection of the Europa-1/South America-1 lineage corresponded to P3: 5'-ACCTTGCTTGCTATCACTGG-3' and to P4: 5'-GCACCATCCAGGT TGCTATT-3', which allowed the amplification of a fragment of DNA of about 940 base pairs (Annex 3).
Samples
For the analysis, 20 samples of viral RNA positive to the N gene of the CDV of national isolates were used without knowing the lineage to which they belonged, which were stored at -20° C in the Virology and Microbiology laboratories of the Faculty of Veterinary Sciences and Cattle of the University of Chile.
As a positive control for the RT-PCR reaction, RNA extracted from two positive samples was used to detect the CDV H gene by RT-PCR [22] and subsequently positive to RT-PCR for the detection of the N gene [20]. One of these samples was sequenced and identified within the America-1 lineage, while the other was sequenced and identified within the Europe-1 / South America-1 lineage [22].
As a control for the multiple detection of both primers, a mixture of RNA from the positive controls was used for the America-1 and Europe-1 / South America-1 lineages mentioned above. As a negative control, RNA extracted from blood with anticoagulant (EDTA) was used from two dogs with no risk history and clinical signs of the disease, one without vaccination and the other with vaccination schedule per day, negative to RT-PCR on the gene N. Nuclease-free water was used as reagent control.
Implementation of the RT-PCR technique
Thermal cycler. To perform the RT-PCR technique, an Apollo (CLP, USA) thermocycler with 96 wells of 0.2 mL was used. The RT-PCR protocol is detailed below.
Reverse transcription. We used 25 μL of the SuperScript one step RT-PCR kit with Platinum Taq DNA Polymerase" (Taq DNA polymerase, MgCl2 and deoxyribonucleotide triphosphate), 5 μL of the template RNA and 5 μL of each specific primer (0.1 – 0.2 μM), obtaining a final volume of 50 μL. After introducing the mixture in the thermal cycler, the reverse transcription step was performed according to the manufacturer's guidelines, which were a first stage at 45° C for 30 minutes for the performance of the reverse transcriptase and then a stage at 94° C for 2 minutes for the denaturation of the enzyme.
DNA amplification. The PCR technique contemplates a stage of DNA denaturation, followed by a stage of alignment of the starters and a final stage of elongation to finish a cycle. The DNA denaturation phase was carried out at 94° C for 30 seconds. During the alignment phase, a single temperature of 53° C was used for 30 seconds, determined according to the melting temperature of both pairs of starters (approximately 58° C). The elongation phase contemplated a temperature of 72° C for one minute. After 40 cycles, we proceeded to a final extension stage at 72° C for 8 minutes and then, we continued with the visualization of the amplified product using 2% agarose gel electrophoresis in TAE buffer. For corroboration of the expected amplicon sizes we used AccuRuler 100bp Plus DNA Ladder (Maestrogen®). Electrophoresis was carried out at 90 volts for 45 minutes and then, the gel was incubated with ethidium bromide (0.5 μg/mL) for 30 minutes, to visualize the DNA bands in an ultraviolet light transilluminator and subsequently photographed.
Results
The entry of the nucleotide sequences defined by the access numbers (Annex 2) in the Clustal Ω program allowed to obtain common zones (Table 1, areas with an asterisk) for both the America-1 lineage and the Europe-1 lineage. / South America-1 of the VDC H gene.
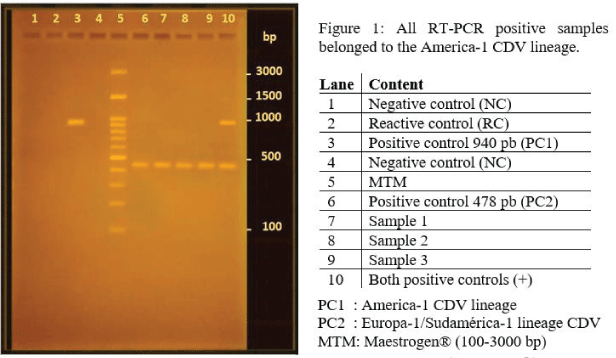
To design primers for the Europe-1/South America-1 lineage, the consensus sequences entered into NCBI's First-BLAST online program were those that presented more than 30 nucleotides in common and a difference with the South America-2 lineage of at least 25 nucleotides. Then, the sequences obtained were compared with the other component sequences of each lineage of the VDC H gene, in addition to being entered the BLAST online program. This strategy helped us confirm that the sequences for each pair of primers were specific to the lineage for which they were designed. The nucleotide sequences with respect to the lineage América-1 and defining an amplicon of 478 bp corresponded to: P1: 5'-CTATTGCATCGGCAGCAAATC-3' and P2: 5'-CCCAGTGTCACTACT AGAATACC-3'. While the sequences to be synthesized regarding the Europe-1/South America-1 lineage and originating an amplicon of 940 bp were: P3: 5'-ACCTTGCTTGCTATCACTGG-3 'and P4: 5'-GCACCATCCAGGTTGCTATT - 3'. In Figure 1 it is possible to observe the bands originated in the RT-PCR reaction of the controls, where clear bands were observed in the case of the positive controls for each of the lineages. No presence of bands was observed in the negative controls or in the reagent control (i.e.: nuclease free water). A molecular size marker between 100 and 3000 bp was used as reference.
All the samples analyzed reacted only with the pair of primers designed for the lineage America-1 (Figures 1,2).
Discussion
CDV remains a common problem, not only in veterinary practice, but also in the conservation of endangered species. It is a highly prevalent disease, very contagious and with many susceptible hosts. Furthermore, although there are vaccines to protect animals from the disease, CDV outbreaks are still observed in vaccinated dogs. It should be mentioned that the vaccines used in Chile are prepared with viral strains belonging to the lineage America-1, one of the two lineages described in the country. According to the literature consulted, there are no epidemiological studies on the prevalence of the Europe-1 / South America-1 lineage in Chile, which would be interesting to explore since an important fraction of cases in domestic dogs vaccinated with CDV are due to a viral strain belonging to the Europe-1/South America-1 lineage.
Because of the analysis of the samples, it was observed that the 20 samples analyzed were positive for the America-1 lineage. These were frozen and dated from 2011, so it would be interesting to continue analyzing other more current samples, to know with greater certainty the prevalence of the Europe-1/South America-1 lineage in the country.
The sample used in this study as a positive control for the Europe-1/South America-1 lineage corresponds to a peripheral blood sample obtained from a symptomatic dog confirmed infected with CDV by ELISA test detecting CDV-specific IgM antibodies. This was further subjected to RT-PCR for detection of the CDV H gene, was sequenced, aligned and segregated within the Europe-1 / South America-1 lineage. The patient was a mestizo female of approximately 1 year, which had vaccination against CDV and presented a neurological presentation of the disease during the year 2011. It would be necessary to collect more information about this patient, such as its travel history, which may help understand its contagion with the Europe-1/South America-1 lineage. In this context, it would also be interesting to know the routine management that was carried out, for example, with the management of secretions. Thus, one might think that if good care was maintained, this viral strain could not spread further and the discovery of this lineage in the country was incidental.
At present, the RT-PCR technique for the H gene is not considered the best diagnostic alternative for the disease due to its high genetic heterogeneity. However, the diagnosis of the different lineages must be carried out based on the analysis of the H gene, and it is optimal that the sample to be analyzed is known to be positive to the CDV. The present study determined that for the established conditions, the design of the primers for each lineage was effective, so it could be suggested as a useful diagnostic tool for epidemiological studies.
In this work we tried to design the most suitable primers for each lineage, which would be exclusive and selective for each one, considering the characteristics that they should have, to be used in the multiple RT-PCR technique. However, there are many similarities among some lineages, as is the case of the Europe-1 / South America-1 and South America-2 lineages. During the design of the oligos described here, a comparison stage of the primers obtained with other lineages was considered. For this reason, it would be interesting to obtain a sample of other CDV lineages, such as, for example, the South America-2 lineage, in order to verify the total selectivity of the primers for each lineage. It should be mentioned that during the RT-PCR technique, the alignment phase of the primers was carried out at a single temperature and the results were optimal with no further need of applying a gradient of annealing temperatures. Finally, it would be interesting to know about the existence of a recently described phylogenetic tree, and together with this literature support, to be able to establish updated nucleotide identity regions.
Conclusion
According to the results obtained after the established RT-PCR protocol, it was observed that the designed primers were able to adequately recognize their relevant lineage with high specificity. The analysis of the samples indicated that all were positive for the America-1 lineage, suggesting that it is the predominant lineage circulating in the country. A larger survey of clinical isolates of CDV would be required to confirm this observation and to carry out epidemiological studies to determine the real prevalence of the Europe-1/South America-1 lineage in Chile at present. This work has resulted in a technique capable of supporting these epidemiological studies.
References
- Vito Martella DVM, Gabrielle Elia DVM, Canio Buonavoglia DVM. Canine Distemper Virus. Vet Clin North Am Small Anim Pract. 2008; 38: 787-797. Ref.: https://goo.gl/knwvHW
- Maclachlan N, Dubovi EJ. Paramyxoviridae. In: Fenner’s Veterinary Virology. 4ª edición. Academic Press. Londres Inglaterra. 2011; 299–325. Ref.: https://goo.gl/n9VN2R
- International Committee on Taxonomy of Viruses. Virus Taxonomy. 2017; Ref.: https://goo.gl/E5PGXZ
- Sarute N, Calderón MG, Pérez R, La Torre J, Hernández M, et al. The fusion protein signal-peptide-coding region of canine distemper virus: a useful tool for phylogenetic reconstruction and lineage identification. PLOS One. 2013; 8: 1-6. Ref.: https://goo.gl/Q5qWmV
- Panzera Y, Calderón MG, Sarute N, Guasco S, Cardeillac A, et al. Evidence of two co-circulating genetic lineages of canine distemper virus in South America. Virus Res. 2012; 163: 401-404. Ref.: https://goo.gl/CPXcht
- Sarute N, Pérez R, Aldaz J, Alfieri AA, Alfieri AF, et al. Molecular typing of canine distemper virus strains reveals the presence of a new genetic variant in South America. Virus Genes. 2014; 48: 474-478. Ref.: https://goo.gl/FkLncF
- Bi Z, Wang Y, Wang X, Xia X. Phylogenetic analysis of canine distemper virus in domestic dogs in Nanging, China. Arch Virol 160: 523-527. Ref.: https://goo.gl/bnhL2g
- Fischer CDB, Gräf T, Ikuta N, Lehmann FKM, Passos DT et al. Phylogenetic analysis of canine distemper virus in South America clade 1 reveals unique molecular signatures of the local epidemic. Infect Genet Evol. 2016; 41:135-141. Ref.: https://goo.gl/ih2hNR
- Beineke A, Puff C, Seehusen F, Baumgärtner W. Pathogenesis and immunopathology of systemic and nervous canine distemper. Vet Immunol Immunopathol. 2009; 127: 1-18. Ref.: https://goo.gl/BpFMLZ
- Espinal MA, Díaz FJ, Ruiz-Saenz J. Phylogenetic evidence of a new canine distemper virus lineage among domestic dogs in Colombia, South America. Vet Microbiol. 2014; 172: 168-176. Ref.: https://goo.gl/RVzAts
- Beineke A, Baumgärtner W, Wohlsein P. Cross-species transmission of canine distemper virus – an update. One Health. 2015; 1: 49-59. Ref.: https://goo.gl/svn7Sp
- Moreira R, Stutzin M. Estudio de la Mortalidad de Zorros en la IV Región. Boletín Veterinario Oficial N°3. Servicio Agrícola Ganadero, División de Protección Pecuaria, Chile. 2005; 1-8. Ref.: https://goo.gl/gHmeuh
- Jara C, Matus P, Moreira R. Distemper canino en Isla Robinson Crusoe (Archipiélago Juan Fernández, V Región): antecedentes de un brote epidémico, 2007. Boletín Veterinario Oficial N°8. Servicio Agrícola Ganadero, División de Protección Pecuaria, Chile. 2007; 1-19. Ref.: https://goo.gl/wZfGWR
- Berríos P, Pincheira B. Distemper canino y su impacto en la fauna silvestre. Científica. 2016; 13: 137-148. Ref.: https://goo.gl/RTe6nV
- Sykes J. Canine Distemper Virus Infection. In: Canine and Feline Infectious Diseases. Elsevier Health Sciences. London, England. 2014; 152-165. Ref.: https://goo.gl/GmRz6G
- Zhao J, Shi N, Sun Y, Martella V, Nikolin V, et al. Pathogenesis of canine distemper virus in experimentally infected racoon dogs, foxes, and minks. Antivir Res. 2015; 122: 1-11. Ref.: https://goo.gl/9U1iua
- Pinotti M, Gollan A, Delgado A, Passeggi C, Occhi H, et al. Distemper canino. Fave. 2009; 8: 29-45. Ref.: https://goo.gl/v9D1ry
- Castilho J, Brandão P, Carnieli P, Oliveira R, Macedo C, et al. Molecular analysis of the N gene of canine Distemper virus in dogs in Brazil. Arq Bras de Med Vet Zootec. 2007; 59: 654-659. Ref.: https://goo.gl/h1dt5z
- Bosco E. Distemper canino. In: Retamal P, Ábalos P, Fredes F. Enfermedades animales producidas por agentes biológicos. Editorial Universitaria. Chile. 2010; 152-156. Ref.: https://goo.gl/9Mp46g
- Muñoz C. Diagnóstico molecular del virus distemper canino mediante la reacción en cadena de la polimerasa asociada a transcripción inversa del gen de la proteína de la nucleocápside viral. Memoria de Título Médico Veterinario. Santiago, Chile. U. Chile, Fac. Medicina Veterinaria. 2013; 32. Ref.: https://goo.gl/RuXfQy
- Ke GM, Ho C, Chiang M, Sanno-Duanda B, Chung C, et al. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet Res. 2015; 11:164. Ref.: https://goo.gl/YybDk2
- Salas V, Pizarro J, Navarro C. Phylogenetic analysis of canine distemper virus detected in Chile. International Journal of Current Research. 2018; 10: 72402-72407. Ref.: https://goo.gl/GG4qTm