More Information
Submitted: November 27, 2020 | Approved: December 10, 2020 | Published: December 11, 2020
How to cite this article: Chae SJ, Lim JS, Lee DY. Characterization of Salmonella spp. isolated from small turtles and human in Republic of Korea. Insights Vet Sci. 2020; 4: 051-055.
DOI: 10.29328/journal.ivs.1001027
Copyright License: © 2020 Chae SJ, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Salmonella infection; Turtles; Zoonoses; Multilocus sequence typing (MLST); Pulsed-field gel electrophoresis (PFGE)
Characterization of Salmonella spp. isolated from small turtles and human in Republic of Korea
Su-Jin Chae1, Jin-Suk Lim2 and Deog-Yong Lee1*
1Division of Viral Diseases, Center for Laboratory Control of Infectious Diseases, Korea Centers for Disease Control and Prevention, Osong, Republic of Korea
2Division of Microbiology, Jeju Institute of Health and Environment, Jeju-do, Republic of Korea
*Address for Correspondence: Deog-Yong Lee (DY Lee). D.V.M., Ph.D., Division of Viral Diseases, Center for Laboratory Control of Infectious Diseases, Korea Centers for Disease Control and Prevention. 187 Osongsaengmyeong 2(i)-ro, Osong-eup, Heungdeok-gu, Cheongju-si, Chungcheongbuk-do, 28159, Republic of Korea, Tel: +82-43-719-8194; Fax: +82-43-719-8219; Email: [email protected]
In 2013, the World Health Organization (WHO) reported that small, pet turtles had caused multistate Salmonella outbreaks in the United States, from where small turtles were subsequently exported into the Republic of Korea. We investigated cases of salmonellosis in South Korea associated with domestic small turtles and analysed genetic characteristics of Salmonella isolates in commercially-available small turtles. We traced six Salmonella serovars, known to have caused human infection in the United States (S. Sandiego, S. Pomona, S. Poona, S. Newport, I 4,(5),12:i:-, and S. Typhimurium), in isolates from suspected Salmonella infection cases in Korea from 2006 to 2015. Additionally, we conducted a pilot study of isolates from small turtles being sold in Korean markets, and performed molecular genetic analysis on the identified strains. S. Pomona was identified in one Salmonella infection case, while all strains isolated from small turtles belonged to either subspecies I (enterica, n = 10, 71.4%) or subspecies IIIb (diarizonae, n = 4, 28.6%). Two serovars (S. Pomona and S. Sandiego) that were highly associated with turtle-to-human transmission were identified with 100% homology to human isolates. Previous to this study, turtle-associated human S. Pomona infections were not well reported in Korea. We report Salmonella infection in small turtles in Korea, and confirm that small turtles should be considered the first infectious agent in S. Pomona infection. We therefore suggest quarantine measures for importing small turtles be enhanced in Korea.
Salmonella spp. is a zoonotic pathogen often reported in humans where there was exposure to infected pets and livestock products [1-3]. Salmonella colonize the intestine of turtles, which are the main source of human infection due to shedding of bacteria in faeces [4]. In the United States, 15 outbreaks of Salmonella associated with small turtles were reported between 2006 and 2014 [5], with most patients exposed to small turtles (less than 4 inches long) raised on two farms in Louisiana. Thirty one percent of the patients were children under 10 years, of which 70% were under 1 year. In 2013, in accordance with the International Health Regulations, the World Health Organization (WHO) noticed the risk of Salmonella infections to 20 countries, including South Korea that imported turtles from the United States. Therefore, we conducted an epidemiological and microbiological survey on the risk of Salmonella infection from imported small turtles in Korea.
Case study on human Salmonella infection
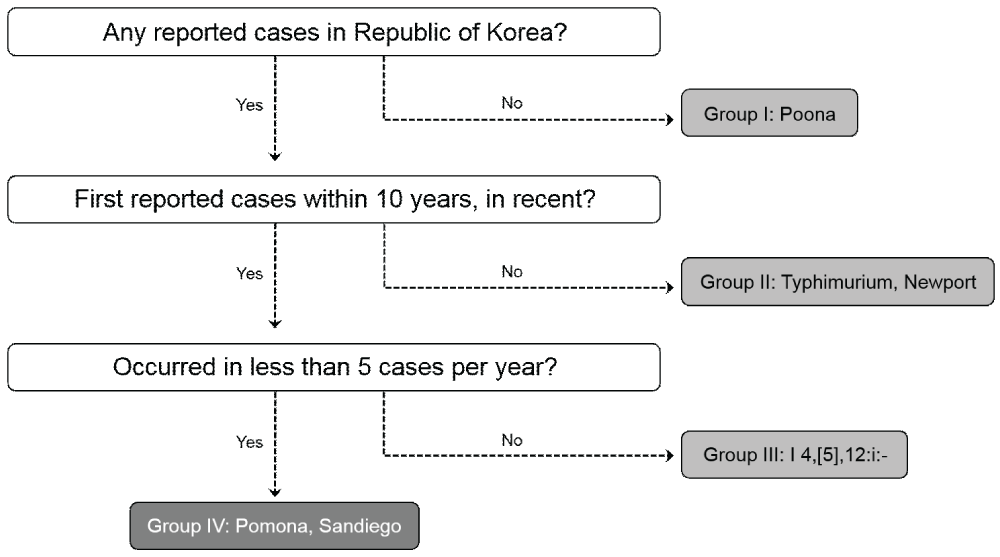
We examined six Salmonella serovars (S. Sandiego, S. Pomona, S. Poona, S. Newport, S. I 4, [5], 12:i:-, and S. Typhimurium) to investigate the possibility of Salmonella spp. infections through small turtle in Korea. We analysed the prevalence of the serovars in isolates from Salmonella infection cases in Korea during the last ten year. The six serovars were classified for inclusion in the epidemiological study according to three criteria: 1) the serovar was not reported in Korea before, 2) the serovar was only reported in the last decade, and 3) less than five isolates of the serovar were reported per year. Serovars not fitting any of the criteria were deemed candidates most likely transmitted from small turtles, and the epidemiological survey was then conducted on these.
Samples from domestic turtles
Five small turtles were purchased from markets in four randomly selected regions of Korea. Turtles were bred over 5 weeks in separate tanks to prevent cross-contamination, and breeding water was changed every 2-3 weeks. Samples from breeding water were collected weekly to analyse the colonisation of Salmonella in the turtles’ intestines. In addition, samples of breeding water from turtles on Jeju Island (Jeju-si, Seogwipo-si) were collected weekly, as those turtles were associated with suspected human infection previously
Isolation and identification of Salmonella spp. strains
Isolation and identification of Salmonella strains was performed following the Korea Centers for Disease Control and Prevention (KCDC) laboratory guidelines. 1 mL breeding water was inoculated into 9 mL Selenite Cysteine Broth (BD Diagnostic Systems™, Sparks, MD, US). The inoculum was then incubated overnight and a loopful of sample plated on MacConkey agar, Salmonella-Shigella agar, and Xylose Lysine Deoxycholate agar (BD™). The unique colonies were streaked on Kligler Iron agar (BD™), and their identification was performed using a biochemical test with API 20E Kit (Biomerieux™, Marcy-1 Etoile, France) and serological testing done as per Lee, et al. [6]. In addition, invA specific PCR was performed to confirm the microbiological identity [7]. Genomic DNA was extracted using the boiling method [8] and amplified by singleplex PCR with specific primers for the invasion of Salmonella into eukaryotic host cells (invA F; 5´-ATT AAT TAT GGA AGC GCT CGC ATT-3´, invA R; 5´-GTA ATG AGA TCC ATC AAA TTA GCG-3´). Serovars were determined by the combination of somatic O and flagellar H antigens according to the Kauffmann-White classification scheme [9,10].
PFGE, MLST & antimicrobial susceptibility of Salmonella strains
To investigae the epidemiological relationship, S. Pomona and S. Sandiego isolates (Group IV in our classification) were analysed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) to compare with human isolates. PFGE was performed according to the PulseNet standard protocol (http://www.pulsenetinternational.org/protocols/). DNA of each isolate was digested with XbaI and BlnI restriction enzymes and separated by CHEF Mapper XA system (Bio-Rad, CA, USA). Salmonella Braenderup (ATCC BAA664) was used as a reference marker strain. PFGE patterns were analysed using BioNumerics v5.1 software (Applied Maths, Saint-Martens-Lartem, Belgium). The isolates were subjected to MLST by determination of the sequences for seven housekeeping genes: aroC, dnaN, hemD, hisD, purE, sucA, and thrA [11]. Sequencing of the nucleotides was performed with an automated sequencer (ABIPRISM 3730XL, Foster City, CA, USA), and the sequence type was determined using analysis tools on the MLST website (http://mlst.warwick.ac.uk/mlst).
An antimicrobial susceptibility test was performed using the VITEK 2 system with AST-N169 card (Biomérieux, Marcy-1´Etoile, France). Susceptibility of the following antimicrobial agents was tested: ampicillin, amoxicillin-clavulanic acid, amikacin, chloramphenicol, cephalothin, ciprofloxacin, ceftriaxone, cefotetan, cefotaxime, cefazolin, cefoxitin, gentamicin, imipenem, nalidixic acid, ampicillin-sulbactam, trimethoprim-sulfamethoxazole, and tetracycline. The Escherichia coli ATCC 25922 card was used for quality control. Results were interpreted as recommended by CLSI guidelines [12].
Case study on human Salmonella infection
A total 5,867 Salmonella isolates were collected from diarrheal patients in Korea from 2006 to 2015 (Table 1). A total of 1,144 strains were identified belonging to six serovars which were thought to be related with small turtles, and were divided into four groups according to classification criteria (Figure 1). No isolates of Group I (Gr. I), S. Pomona, have been reported in Korea, and this serovar was therefore excluded from the case study. Group II, (Gr. II) including the S. Typhimurium (n = 745) and S. Newport (n = 47) serovars, were excluded from the study as there were too many cases to link logistically to small turtles. Group III (Gr. III) isolates, from the Salmonella I 4,[5],12:i:-, serovar (n = 346), were also excluded; cases of this serotype, a variant strain of S. Typhimurium, have been continuously increasing since it was first reported in 2008. In addition, Gr. II and Gr. III isolates were excluded for the variety of host ranges. Therefore, Group IV (Gr. IV) isolates, S. Sandiego (n = 4) and S. Pomona (n = 2) serovars, were remaining as possible candidates to link with transmission from turtles; isolates of these serotypes have been reported separately within the last five years, with no more than five isolates annually. However, were able to identify just a single suspected case associated with S. Pamona.
| Table 1: Prevalence of Salmonella serovars associated with small turtles imported from the USA to Korea, 2006-2015. | ||||||||
| Number of organisms | Serovars | |||||||
| Gr. I | Gr. II | Gr. III | Gr. IV | Others | Total Salmonella spp. |
|||
| Poona | Newport | Typhimurium | I 4,[5],12:i:- | Pomona | Sandiego | |||
| 2006 | 0 | 1 | 38 | 0 | 0 | 0 | 332 | 371 |
| 2007 | 0 | 3 | 30 | 0 | 0 | 0 | 390 | 423 |
| 2008 | 0 | 4 | 49 | 11 | 0 | 0 | 306 | 370 |
| 2009 | 0 | 0 | 109 | 4 | 0 | 0 | 394 | 507 |
| 2010 | 0 | 2 | 72 | 18 | 0 | 0 | 605 | 697 |
| 2011 | 0 | 6 | 133 | 30 | 0 | 1 | 799 | 969 |
| 2012 | 0 | 5 | 62 | 30 | 0 | 2 | 266 | 365 |
| 2013 | 0 | 16 | 101 | 101 | 2 | 0 | 520 | 740 |
| 2014 | 0 | 3 | 99 | 77 | 0 | 1 | 674 | 854 |
| 2015 | 0 | 7 | 52 | 75 | 0 | 0 | 437 | 571 |
| Total | 0 | 47 | 745 | 346 | 2 | 4 | 4723 | 5,867 |
A case of S. Pomona infection in Korea
A two-year-old boy began to show symptoms of fever on July, 7. 2013. He visited the emergency room two days later with symptoms of fever, abdominal pain, and diarrhoea. The patient returned home after receiving fluid therapy in combination with antispasmodic, sedative, anti-inflammatory, and analgesic therapies. Soon after, he visited the nearby paediatric and juvenile clinic, displaying the symptoms of fever, abdominal pain, and watery diarrhoea again, along with vomiting, poor oral intake, and weight loss (14.8-16 kg). In the evening, he was re-admitted to the emergency room and received intravenous UBACSIN treatment (Jeilpharm, Korea). He remained febrile and had to be hospitalized. Within two days of hospitalization, treatment was changed to cefotaxime and symptoms improved. In microbiological examination, non-typhoidal Salmonella was isolated from patient’s faecal samples and serotyping identified S. Pomona (M group, 28:y,1,7). During the epidemiological investigation, the patient’s parents reported that the child had begun to experience symptoms after touching a turtle while visiting a neighbour. Samples from the neighbour’s turtle could not be analyzed as the owner refused to allow testing on the animal, but we purchased two turtles from a nearby traditional market to investigate Salmonella infection in turtles in the relevant area.
Isolation and identification of Salmonella spp. from domestic turtles
A total 14 Salmonella strains were isolated from five small turtles purchased at randomly chosen commercial markets and two turtles purchased at a traditional market in an area where a patient may have contracted Salmonella from a domestic turtle. Ten of the 14 strains belonged to subspecies enterica and the remaining four strains belonged to subspecies diarizonae. Seven serovars were identified, of which the most frequent was S. Pomona (n = 5), followed by S. Litchfield (n = 2), S. Montevideo (n = 2), S. Sandiego (n = 1), S. IIIb 50:k:z (n = 1), S. IIIb 60:[k]:z53 (n = 2), and S. IIIb 61:c:1,5,[7] (n = 1) (Table 2).
| Table 2: Distribution of Salmonella subspecies and serovar strains isolated in breeding water of small turtles. | ||||||||
| Salmonella subsp. Serovar |
Samples | No. of isolates | ||||||
| A | B | C | D | E | JJ1* | JJ2* | ||
| Salmonella enterica | ||||||||
| Litchfield | + | - | + | - | - | - | - | 2 |
| Montevideo | + | - | + | - | - | - | - | 2 |
| Pomona | + | - | + | + | + | + | - | 5 |
| Sandiego | - | - | - | - | - | - | + | 1 |
| Salmonella diarizonae | ||||||||
| IIIb 50:k:z | - | - | - | - | + | - | - | 1 |
| IIIb 60:[k]:z53 | + | - | + | - | - | - | - | 2 |
| IIIb 61:c:1,5,[7] | - | - | - | - | - | - | + | 1 |
PFGE, MLST, and antimicrobial susceptibility testing
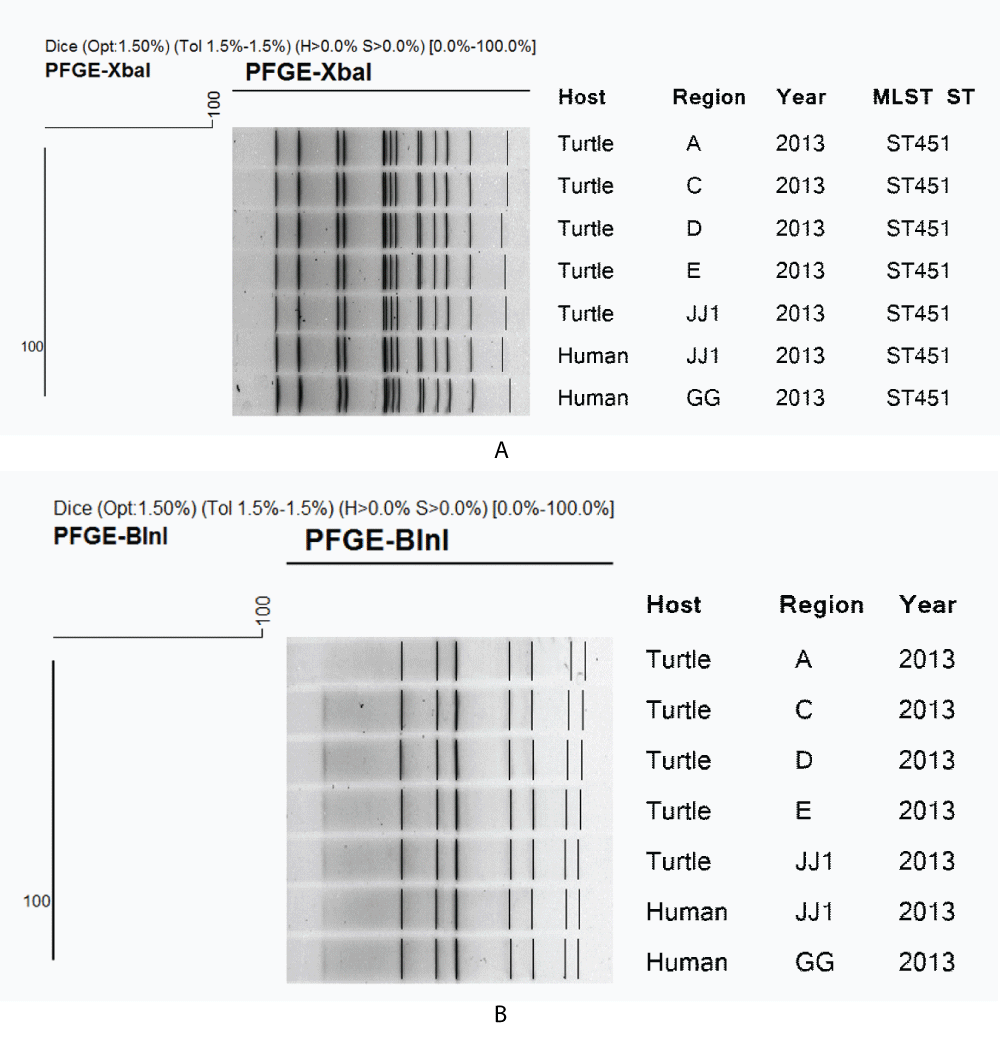
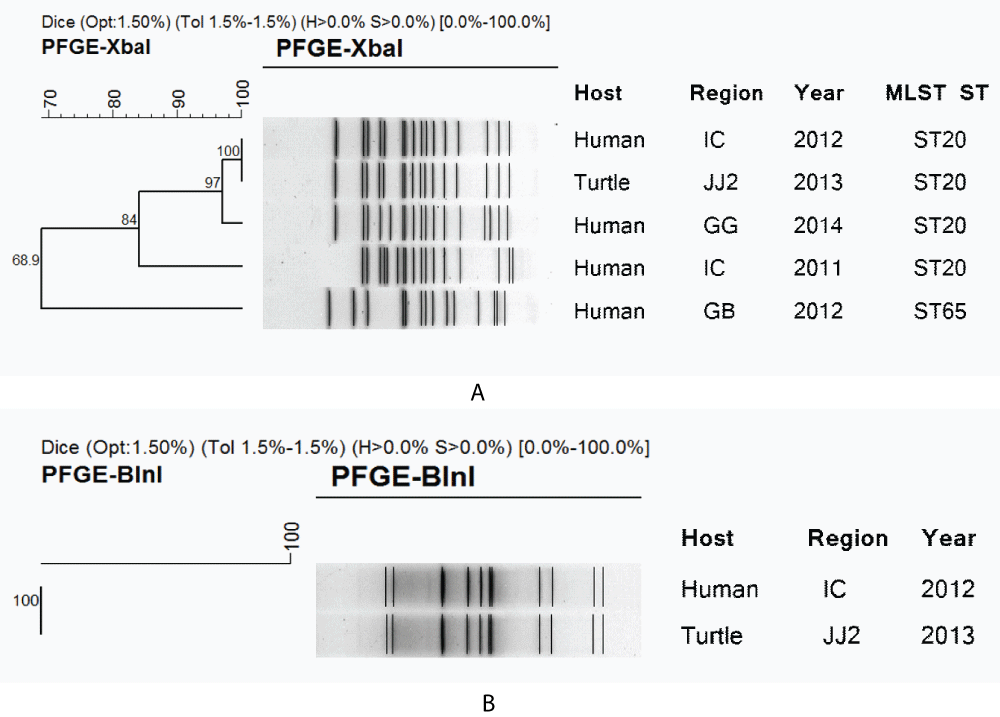
We conducted PFGE and MLST for the characterization of S. Pomona and S. Sandiego which were commonly identified through risk assessment and bacterial isolation from turtles. For S. Pomona, human (n = 2) and turtle isolates (n = 5) showed 100% homology on PFGE, while MLST showed the same sequence type as the ST451 strain (Figure 2). In contrast, S. Sandiego was divided into four clusters as a result of PFGE performed on human (n = 4) and turtle isolates (n = 1). However, human strains (year 2012) and suspected turtle-related strains showed 100% homology on PFGE and the same sequence type as the ST20 strain on MLST (Figure 3). All Salmonella isolates from turtles and humans were susceptible to the 17 antimicrobial agents tested.
Figure 1: Classification of Salmonella serovars associated with small turtles imported from the USA to Korea, 2006-2015.
Figure 2: Clustering of (A) XbaI and (B) BlnI–digested DNA fragments by pulsed-field gel electrophoresis (PFGE) with multilocus sequence typing (MLST) for Salmonella Pomona strains isolated from turtles and human cases. A, B, C, and D regions are randomized and JJ and GG are abbreviations of Jeju Island and Gyeonggi Province, respectively.
Figure 3: Clustering of (A) XbaI and (B) BlnI–digested DNA fragments by pulsed-field gel electrophoresis (PFGE) with multilocus sequence typing (MLST) sequence type for Salmonella Sandiego strains isolated from turtles and human cases. IC, JJ, and GG are abbreviations of Incheon Metropolitan City, Jeju Island, and Gyeonggi Province, respectively.
Salmonella enterica subspecies enterica (I) are mainly isolated from warm-blooded animals; while other subspecies (II, IIIa, IIIb, IV and VI) and Salmonella bongori (V) are mainly isolated from cold-blooded animals and from the environment. However, most isolates from turtles in this study were identified as subsp. enterica. In recent years, subsp. enterica has been identified in a large number of Salmonella infection cases associated with reptiles [13-15]. Generally, turtles carry Salmonella spp. in their intestinal tracts, and continuously release the bacteria through faeces, causing human infection indirectly. Salmonella, which is asymptomatic in reptiles, can cause serious illness in humans. Reptile-associated salmonellosis (RAS) can cause sepsis, meningitis, and bone and joint infections, especially in children [14]. Transmission from turtles accounts for 42% of RAS infections, and children are more affected because they often interact with domestic turtles.
In the United States, at least 473 cases of Salmonella infection due to small turtles were reported from March 2012 through October 2013 [16]. The cases were linked to turtles raised on two farms in Louisiana and exported to more than 20 countries. According to statistics from the Korea Customs Service, 28.76 tons of turtles were imported into the Republic of Korea annually over the past sixteen years, of which 20.17% were imported from the United States [17]. It is therefore concerning that turtles can be imported into Korea without quarantine, making it difficult to conduct epidemiologic studies to link turtles to cases of disease because the animals are often imported directly from the United States on a small scale.
No cases of Salmonella infection caused by small turtles have been reported in Korea as yet. However several papers reported a possible case of suspected transmission from turtles [18] with cases of rare serovars [19,20]. In the case identified here, it was epidemiologically certain that the patient was infected through contact with a turtle, but it was not confirmed experimentally. Additionally, the identified serovar, S. Pomona was genetically identical in isolates from the patient and turtles purchased from a regional traditional market. Moreover, S. Pomona strains isolated from additional local patients and further local small turtles were genetically identical to the patient’s strain. The case is also similar to two cases previously reported in children younger than 5 years who required treatment [21,22], and we suggest this is the first confirmed case of Salmonella infection through turtle in Korea.
Antimicrobial agents are used to reduce Salmonella infection in reptiles, which may cause antibiotic resistance [23]. Salmonella isolates from reptiles, especially from turtles, were identified to exhibit a variety of multidrug resistance [24]. All isolates in this study were susceptible to the 17 agents used and no antimicrobial resistance genes were detected. However, ampicillin-sulbactam treatment against S. Pomona in the identified case did not relieve clinical symptoms, and symptoms only improved after treatment was changed to the cephalosporin, cefotaxime, similar to Kim, et al. [18] who recommended that a third-generation cephalosporin be used in cases of suspected non-typhoidal Salmonella bacteraemia with resistance to ampicillin. This may mean experience-based antimicrobial therapy is effective when Salmonella is transmitted from small turtle to human.
Small turtles available in the Korean domestic market have Salmonella in their intestines. Although Salmonella infection does not often cause serious illness, high-risk groups such as the immune-compromised, children under 5 years, and pregnant women are advised to avoid contact with small turtles. In addition, quarantine of exotic pets such as turtles should be strengthened as domestic imports continue to increase.
This study was supported by Acute Diarrheal Laboratory Surveillance (EnterNet-Korea) in Korea (4800-4851-304).
Ethics statement
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to. No ethical approval was required.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors’ contributions
Su-Jin Chae was responsible for writing the original draft. Jin-Suk Lim was responsible for the methodology and investigation. Deog-Yong Lee was responsible for the conceptualization and supervision of the study.
- CDC. Salmonellosis associated with pet turtles--Wisconsin and Wyoming, 2004. MMWR. 2005; 54. 223-226. PubMed: https://pubmed.ncbi.nlm.nih.gov/15758895/
- Seepersadsingh N, Adesiyun AA. Prevalence and antimicrobial resistance of Salmonella spp. in pet mammals, reptiles, fish aquarium water, and birds in Trinidad. J Vet Med B Infect Dis Vet Public Health. 2003; 50: 488-493. PubMed: https://pubmed.ncbi.nlm.nih.gov/14720186/
- Woodward DL, Khakhria R, Johnson WM. Human salmonellosis associated with exotic pets. J Clin Microbiol. 1997; 35: 2786-2790. PubMed: https://pubmed.ncbi.nlm.nih.gov/9350734/
- DuPonte MW, Nakamura RM, Chang EM. Activation of latent Salmonella and Arizona organisms by dehydration of red-eared turtles, Pseudemys scripta-elegans. Am J Vet Re. 1978; 39: 529-530. PubMed: https://pubmed.ncbi.nlm.nih.gov/637401/
- Bosch S, Tauxe RV, Behravesh CB. Turtle-Associated Salmonellosis, United States, 2006-2014. Emerg Infect Dis. 2016; 22: 1149-1155. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4918145/
- Lee DY, Lee E, Min JE, Kim SH, Oh HB, et al. Epidemic by Salmonella I 4,[5], 12: i:-and characteristics of isolates in Korea. J Infection Chemotherapy. 2011; 43:186-190.
- Kim SH, Kim SH, Lee SW, Kang YH, Lee BK. Rapid serological identification for monophasic Salmonella serovars with a hin gene: specific polymerase chain reaction. J Bacteriol Virol. 2005; 35: 291-298.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual: Cold spring harbor laboratory press; 1989.
- Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars. WHO collaborating centre for reference on Salmonella. 2007; 9: 1-166.
- Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, et al. Supplement 2008–2010 (no. 48) to the white–Kauffmann–Le minor scheme. J Res Microbiol. 2014; 165: 526-530. PubMed: https://pubmed.ncbi.nlm.nih.gov/25049166/
- Yun YS, Chae SJ, Na HY, Chung GT, Yoo CK, et al. Modified method of multilocus sequence typing (MLST) for serotyping in Salmonella species. J Bacteriol Virol. 2015; 45: 314-318.
- Institute CaLS. Performance standards for antimicrobial susceptibility testing. 2018; 23.
- Geue L, Loschner U. Salmonella enterica in reptiles of German and Austrian origin. Vet Microbiol. 2002; 84: 79-91. PubMed: https://pubmed.ncbi.nlm.nih.gov/11731161/
- Meyer Sauteur PM, Relly C, Hug M, Wittenbrink MM, Berger C. Risk factors for invAsive reptile-associated salmonellosis in children. Vector Borne Zoonotic Dis. 2013; 13: 419-421. https://pubmed.ncbi.nlm.nih.gov/23473215/
- Nakadai A, Kuroki T, Kato Y, Suzuki R, Yamai S, Yaginuma C, et al. Prevalence of Salmonella spp. in pet reptiles in Japan. J Vet Med Sci. 2005; 67: 97-101. PubMed: https://pubmed.ncbi.nlm.nih.gov/15699603/
- Khuly P, Meyer T, Carlson R, Delamaide Gasper JA, Rylander H, et al. Eight multistate outbreaks of human Salmonella infections linked to small turtles (final update). J Am Vet Med Assoc. 2014; 244: 1243-1245.
- KCS. Trade statics system. Korea Custom Service. 2018. http://www.customs.go.kr.
- Kim JD, Choi SY, Kim DS, Kim KH. Two Children with Nontyphoidal Salmonellosis Assumed by Pets Korean J Pediatr Infect Dis. 2013; 20: 41-45.
- Chae SJ, Yun YS, Yoo CK, Chung GT, Lee DY. First Report of Salmonella Serotype Tilene Infection in Korea. J Annals of Clinical Microbiology. 2016; 19: 24-27.
- Lee DY, Kang Y. Human infection of Salmonella matadi in Korea. Yonsei Med J. 2013; 54: 1297-1298. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3743207/
- CDC. Turtle-associated salmonellosis in humans--United States, 2006-2007. MMWR. 2007; 56: 649.
- Tauxe RV, Rigau-Perez JG, Wells JG, Blake PA. Turtle-associated salmonellosis in Puerto Rico. Hazards of the global turtle trade. JAMA. 1985; 254: 237-239. PubMed: https://pubmed.ncbi.nlm.nih.gov/3999366/
- Cooke FJ, De Pinna E, Maguire C, Guha S, Pickard DJ, et al. First report of human infection with Salmonella enterica serovar Apapa resulting from exposure to a pet lizard. J Clin Microbiol. 2009; 47: 2672-2674. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5617995/
- Chen CY, Chen WC, Chin SC, Lai YH, Tung KC, et al. Prevalence and antimicrobial susceptibility of Salmonellae isolates from reptiles in Taiwan. J Vet Diagn Invest. 2010; 22: 44-50. PubMed: https://pubmed.ncbi.nlm.nih.gov/20093681/